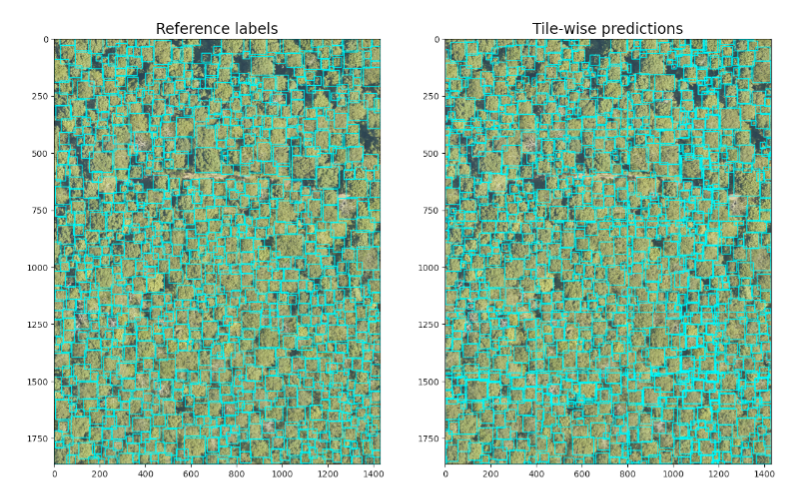
Tree crown detection using DeepForest
How to run¶
Running on Binder¶
The notebook is designed to be launched from Binder.
Click the Launch Binder button at the top level of the repository
Running locally¶
You may also download the notebook from GitHub to run it locally:
Open your terminal
Check your conda install with
conda --version. If you don’t have conda, install it by following these instructions (see here)Clone the repository
git clone https://github.com/eds-book-gallery/15d986da-2d7c-44fb-af71-700494485def.gitMove into the cloned repository
cd 15d986da-2d7c-44fb-af71-700494485defCreate and activate your environment from the
.binder/environment.ymlfileconda env create -f .binder/environment.yml conda activate 15d986da-2d7c-44fb-af71-700494485defLaunch the jupyter interface of your preference, notebook,
jupyter notebookor labjupyter lab
- Coca-Castro, A. (2025). Tree crown detection using DeepForest (Jupyter Notebook) published in the Environmental Data Science book. Zenodo. 10.5281/ZENODO.8309661